Synchrotron X-ray footprinting (XF or XFP) provides a new method for studying macromolecular structures in the native solution state. XFP technique employs broadband hard X-ray beams to generate hydroxyl radicals in solution, thus subsequently covalently modifying amino acid side chains of protein structure and cleaving the phosphodiester backbone of nucleic acids.
High flux of synchrotron X-ray beams produce hydroxyl radicals for X-ray footprinting experiments to study structural information and conformational changes of proteins and nucleic acids. Up to now, XFP beamlines have been implemented to two national synchrotron facilities, the Advanced Light Source (ALS) and the National Synchrotron Light Source (NSLS) in USA. The XFP beamline of NSLS utilizes hard X-rays from a three-pole wiggler (3PW) source, thus delivering high flux at sample in the range of 1x1016 phs/s. Figure 1 illustrates the XFP end station, showing various sample delivering systems. X-ray shutter is employed for exposure of sample solution which is contained in the tube. The duration of X-ray exposure much depends on the velocity of the sample passing the X-ray beam in multi-well plate setup and jet nozzle, or capillary.
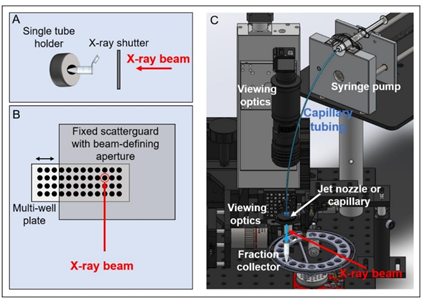
Figure 1 Sample delivery system at the end station of XFP beamlines. (A) Single tube holder (B) Multi-well plate (C) Jet nozzle or capillary (Ralston & Joshua, 2022).
A schematic diagram of protein X-ray footprinting measurements is shown in Figure 2. An X-ray beam exposes to apo and ligand-bound (holo) proteins, thus subsequently digesting with proteases and generating peptides, separating by Mass Spectrometry. The fragmentation pattern is unique to each peptide; therefore MS/MS spectra is employed to identify unmodified and modified peptides/residues within the sample. Peak identification of extracted ion chromatograms is for quantifying the fraction of the unmodified and modified peptides/residues within the sample and determining regions of the protein which are responsible for specific function.
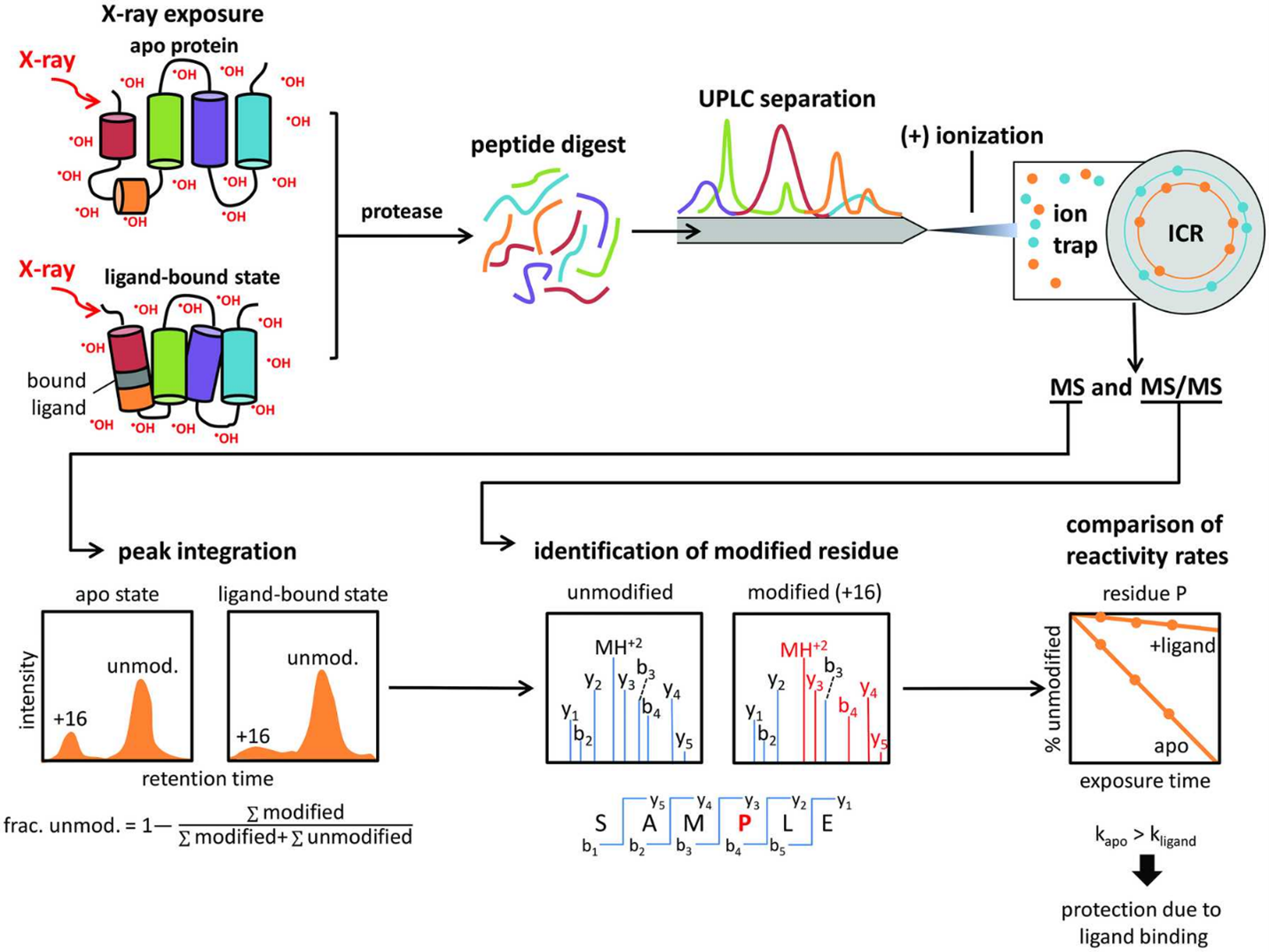
Figure 2. Schematic diagram of protein X-ray footprinting measurement. An X-ray beam exposed to apo and ligand-bound proteins. Digested peptides are separated by ultraperformance liquid chromatography technique (UPLC). Identification of the unmodified and modified peptides/residues is analyzed by electrospray ionization coupled with tandem mass spectrometry (ESI-MS/MS) (Asuru et al., 2019).
Recently, XFP is an ideal complementary technique, thus comparing to low-to-medium resolution data from cryo-EM or solution scattering studies. Although crystallography and NMR provide high-resolution information. However, crystallization procedures and size limitations are much considered as bottlenecks. Static and time-resolved structural analysis of complex structures brings benefit of XFP technique, thus continuously developing to meet its highest performance.
Re-written by Chomphunuch Songsiriritthigul (Beamline Scientist, SLRI)
References
- Asuru, A., Farquhar, E.R., Sullivan, M., Abel, D., Toomey, J., Chance, M.R. & Bohon, J. (2019). The XFP (17-BM) beamline for X-ray footprinting at NSLS-II. Journal of Synchrotron Radiation. 26, 1388-1399. DOI: 10.1107/S1600577519003576.
- Chance, M.R., Farquhar, E.R., Yang, S., Lodowski, D.T. & Kiselar, J. (2020). Protein Footprinting: Auxiliary Engine to Power the Structural Biology Revolution. Journal of Molecular Biology. 432, 2973-2984. DOI: 10.1016/j.jmb.2020.02.011.
- Gupta, S. (2019). Using X-ray Footprinting and Mass Spectrometry to Study the Structure and Function of Membrane Proteins. Protein & Peptide Letters. 26(1):44-54. DOI: 10.2174/0929866526666181128142401.
- Morton, S.A., Gupta, S., Petzold, C.J. & Ralston, C.Y. (2019). Recent Advances in X-Ray Hydroxyl Radical Footprinting at the Advanced Light Source Synchrotron. Protein Peptide Letters. 26(1), 70-75. DOI: 10.2174/0929866526666181128125725.
- Ralston, C.Y. & Joshua S.S. (2022). Structural Investigation of Therapeutic Antibodies Using Hydroxyl Radical Protein Footprinting Methods. Antibodies11. 71. DOI: 10.3390/antib11040071.

